Match Taxa dialog
This dialog enables you to match the taxon names in a data file with those in your database taxon dictionary.
Typical Matching Procedure:
- Press Match All. Any taxa with obvious matches are then linked. This automatic matching works partly on the basis of categories. If there are unrecognised categories in your workspace, you will be asked if you wish to ignore them.
- Select a data source to record matches (see below).
- Match the remaining taxa individually. Double-click on a row (or press Edit/Search) to open the Taxa: Select dialog. Here you can search the database for a similar taxon and match to it (you will need to do this where workspace taxa are spelt incorrectly, for example).
- Exclude any taxa which you either do not wish to import, or cannot match and do not wish to add.
- Any remaining taxa should be genuinely new to your database. You should Add these taxa, either individually from the Taxa: Select dialog, or by pressing Add all.
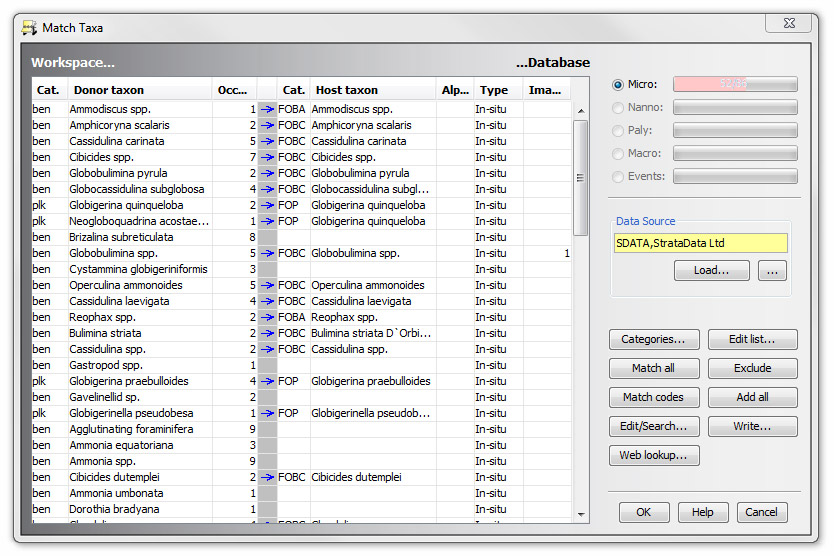
Taxa (and their categories) in the data file (the Donor) are displayed in to the left. Matching taxa in the database (the Host) will be displayed in the right-hand columns. If a match for a particular taxon cannot be found, the relevant database taxon field will be blank. You can translate the unrecognised name to one which is in the database or add it to the database as a new species.
Note: Unmatched taxa often result from spelling and formatting errors - bear this in mind when matching taxa.
The number of occurrences of each taxon in the dataset is displayed in the furthest right hand column of the workspace side. You can use this information to inform decisions about how important it may be to translate a particular taxon name.
You can use the drop-down list in the cells of the Type column to specify that all occurrences in the workspace for that taxon should be updated to the chosen type. The list includes reworked, caved, questionable, and all the sub-types you have set in your database. Use this option where the workspace taxon clearly indicates an occurrence type, e.g. species "Ammonia spp. reworked" could be linked to "Ammonia spp." and the occurrences switched to "Rw". Note that if occurrences already have a sub-type set, and you choose a sub-type from the list, the type you chose will be appended to the existing type to form a new sub-type.
Some database taxa may be linked to images. The number of images of each taxon appears in the right-hand column of the database side. You can open the Image Set dialog by double-clicking in this column where are numbers.
The discipline(s) to which the file's taxa belong are indicated by the progress bars in the top-right corner of the dialog. Where there are no data (as for Nanno in the example), the bars are disabled. Text in the bars indicate how many taxa are matched of the total number that must be matched.
Important: you must complete the match process for each discipline, by selecting each radio button in turn.
Typically you would match as many taxa as possible against your database, and then edit the remaining names which are incorrectly spelt or formatted, and then add genuinely new taxa to the database.
There are a number of options which you select from buttons on the right hand control bar of the dialog as follows:
Data source
A Data Source is an organisation (or individual) from which you receive data.
Note: this is not the same as an analyst. They are effectively the same if an analyst has a personal database, but you also may have data from different analysts who share a common database, or data source.
If you have previously received data from the source that provided the data file you are currently working with, you can select the source by pressing the ellipsis button (...). Any editorial changes you made last time you imported data from this source will then be applied the incoming taxon names in this session. This is useful if you regularly import data from a particular source. If you choose not to select a source, any editorial changes you make will not be stored for future use.
Press Load or the ellipsis (...) in the data source box. Select a data source in the Data Source: Select dialog. You can view past matches from this data source by pressing Matches...
Hint: highlight individual rows or press Select all then press Delete selection to remove matches for this source from the list. You may wish to do this if you previously made a match which you no longer wish to use.
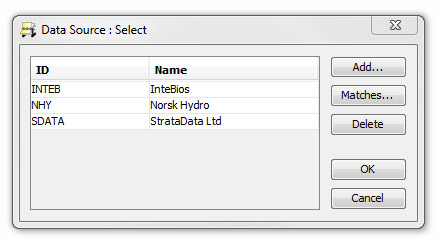
If the data source is new you can also Add it (you will be prompted to enter a description and a name).
Categories
Press Categories... to open the Match Categories dialog.
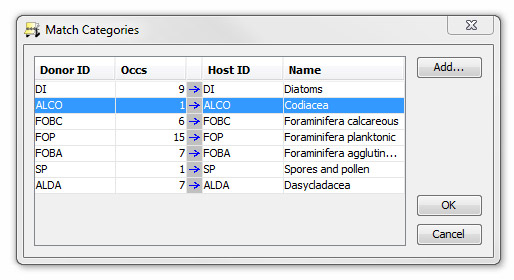
All taxa in StrataBugs must belong to a category. Other users may have assigned taxa to categories differently or added new categories and you may wish to switch some taxa from one category to another.
Categories will be matched automatically where possible. Where unmatched, select an existing category by clicking in the Host ID column.
To add a new category to match to, press Add...
Match all
To match names against all the names in the database press the Match all button. Any names recognised as already present in your StrataBugs database will be added to the Host taxon column on the righthand side of the dialog box. This will not take account of previous matches for the same data source.
Note: If you Load from a previous data source first, those matches will take priority. If you press the Match all button first, taxa will be matched against the database as a priority.
Note: If any taxa have been assigned to Categories in the source data which do not match the ones designated in your database the global search will not retrieve them and you will have to add them individually and assign the correct Category (see Categories above).
Match codes
By default taxa are matched using their full taxonomic names. If you press the Match Codes button, the taxa will be matched by alphanumeric codes. These are convenient abbreviations (e.g. Mel pomp or Ammobac 1) which you may wish to use. There are options in the Chart plotting application to display these taxa either by their full names or alphanumeric codes.
Edit/Search
Use this button where there are taxa which were not matched by the match all function, and you want to:
Search your database for a taxon similar to the donor taxon, which you can then match it to
Select the taxon and press Edit/Search (or double-click on unmatched taxa). The Select Taxa dialog shows, displaying the name of the selected taxon and some suggested search parameters. You may need to make the search parameters more general to get a list of results (as if an exact match existed it would have been found and matched by 'match all').
- Add the donor taxon to your database with or without modifications, which then creates a match
If you know the taxon in your file does not exist in your database, you may wish to add it to the database. Open the Select Taxa dialog as above and press the Add button.
Note: be sure to check the name and the way in which it is formatted before you press OK to add the taxon. Edit any of the fields so that the name is correct. Use the Clear species, Clear genus and Clear all buttons to help edit the name. If the Category is incorrect, select the right one from the drop down menu.
Hint: If you are not sure how to spell or format a name, but you still wish to add it to the database, use the wildcards (%) and press Search to look for a similar name in the database. When you are sure you have the correct name, proceed to add the taxon.
Web lookup
Launches your default browser with a Google search for the full donor taxon name.
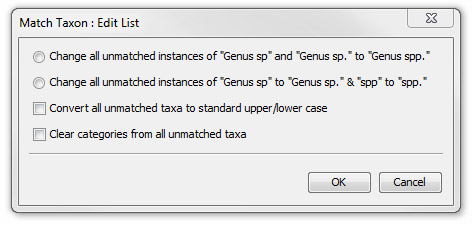
Edit list
To improve the consistency of all unmatched taxon names you may convert "sp" to "spp." or "sp" to "sp." or rectify incorrect capitalisation so that the names will be more likely to match the ones in your database. Press Edit List and select an option form the Match Taxon: Edit List dialog.
Note: Capitalisation will not change any text following the term sp.
Exclude
Highlight one or more taxa on the list and press Exclude to discard them from the import process.
Add all
If you wish to add all the taxa from the source file, irrespective of whether they are matched in the database or not, push the Add all button. This will not overwrite previous selections made using Load or Match all but may add incorrect names to your database.
Note: It is dangerous to do this if you are less than certain about the quality of the data in the data file, particularly spelling and formatting styles. Use this only if you are absolutely certain that all the remaining unmatched names on the list are correct. An example of when this may be advisable is if you are transferring data which you have recorded on one PC to another. Never trust other user's spelling!
Hint: To import a long list of taxa which are not recognised by the database quickly and accurately, scan the list by eye and provide matches for all the names which have more than two parts (i.e. more than genus and species names). Then scan the rest of the list looking for any obvious errors (e.g. sub-species and species epithets together in the same field, cf. and ? included in the species field, etc.). Next use Add all. All the taxa for which the genus name can be recognised are automatically added. Then edit the first species of each remaining genus, providing the correct category assignment and add it. Repeat Add all. Repeat the last steps if there are any left.
Write
This will write out a list of unmatched taxa as a .TXT file.
OK/Cancel
When you press OK , any non-maching taxa will be removed from the analyses in the wells in the workspace. All the taxa in the workspace will be updated to reflect the names assigned from the database. If you Cancel from this dialog, the previous matches will be retained, but the non-matching taxa will not be removed, or database names substituted. You will not be able to proceed to save any well data that does not have a fully matched or filtered set of taxa.
Page last updated: 03-Dec-2014 10:24